
Robust Clustering via NMF (non-negative matrix factorization)
Source:R/nmfClustering.R
nmfClustering.RdWhen one rank is provided, NMF clustering will be performed. When multiple ranks are provided, this function will determine the optimal number of communities (K) by assessing the cophenetic coefficient across different values of K.
Usage
nmfClustering(
mat,
ranks = 10,
nrun.per.rank = 30,
min.coph = 0.95,
nmf.method = "brunet",
ncores = 1,
plot = FALSE,
seed = 2024,
...
)Arguments
- mat
Numeric matrix (feature by sample) for clustering analysis.
- ranks
Numeric vector specifying the number of clusters to evaluate.
- nrun.per.rank
Integer specifying the number of runs per rank for clustering.
- min.coph
Numeric specifying the minimum cophenetic coefficient required for a rank to be optimal.
- nmf.method
Character string specifying the method for NMF analysis.
- ncores
Integer specifying the number of CPU cores to use for parallel processing.
- plot
Logical indicating whether to plot the results.
- seed
An integer used to seed the random number generator for NMF analysis.
- ...
Additional arguments to be passed to the nmf function.
Value
An NMFfitX1 object when only one rank is provided. A list containing the optimal number of communities (bestK), a list of NMFfitX1 objects (NMFfits), and a ggplot object (p) displaying the cophenetic coefficient across different values of K.
Examples
library(NMF)
library(SpatialEcoTyper)
mat <- matrix(rnorm(1000, 3), 20)
mat[mat<0] = 0
## Specify one rank
result <- nmfClustering(mat = mat, ranks = 3, nrun.per.rank = 3)
predict(result)
#> [1] 3 2 2 3 2 2 2 2 3 1 1 1 1 3 1 2 3 3 3 2 2 2 2 1 2 1 3 3 3 3 2 3 1 3 3 2 3 2
#> [39] 1 2 2 3 3 1 3 2 1 3 3 3
#> attr(,"what")
#> [1] columns
#> Levels: 1 2 3
## Determine optimal ranks by testing multiple ranks
result <- nmfClustering(mat = mat, ranks = 2:5, nrun.per.rank = 3)
result$p
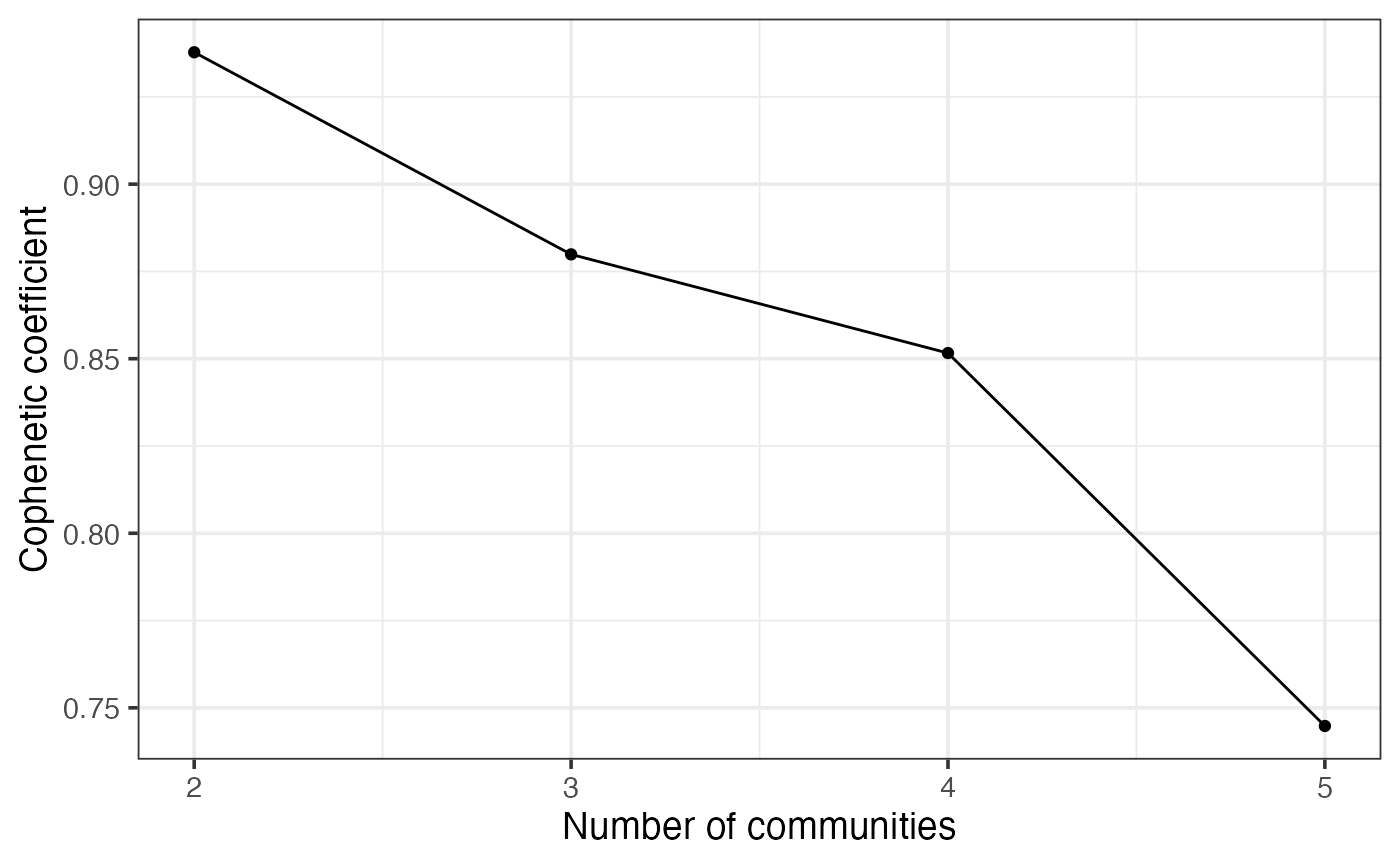 result$bestK
#> [1] 2
predict(result$NMFfits[[paste0("K.", result$bestK)]])
#> [1] 1 1 2 1 1 1 1 1 2 2 2 2 1 2 2 1 1 2 2 1 1 1 2 2 1 2 1 2 1 1 1 2 2 2 1 1 2 1
#> [39] 2 1 2 1 1 2 2 1 2 2 2 2
#> attr(,"what")
#> [1] columns
#> Levels: 1 2
result$bestK
#> [1] 2
predict(result$NMFfits[[paste0("K.", result$bestK)]])
#> [1] 1 1 2 1 1 1 1 1 2 2 2 2 1 2 2 1 1 2 2 1 1 1 2 2 1 2 1 2 1 1 1 2 2 2 1 1 2 1
#> [39] 2 1 2 1 1 2 2 1 2 2 2 2
#> attr(,"what")
#> [1] columns
#> Levels: 1 2